KNN Regression (cont.)
3/7/23
Housekeeping
KNN implementations: due last night to Canvas (please submit your PDF if you haven’t already)
Will grade Lab 02: Moneyball today, and hopefully the KNN implementations tomorrow
Another small KNN regression deliverable due this Thursday 03/09 11:59pm.
K-nearest neighbors: other considerations
Algorithm (recap)
Choose a positive integer \(K\), and have your data split into a train set and test set
For a given test observation with predictor \(x_{0}\):
Identify the \(K\) points in the train data that are closest (in predictor space) to \(x_{0}\). Call this set of neighbors \(\mathcal{N}_{0}\).
Predict \(\hat{y}_{0}\) to be the average of the responses in the neighbor set, i.e. \[\hat{y}_{0} = \frac{1}{K} \sum_{i \in \mathcal{N}_{0}} y_{i}\]
Standardizing predictors
Mite data: preparation
The following code divides my data into train and test sets.
Make sure you understand what each line of code is doing! If you don’t, please ask!
set.seed(6)
n <- nrow(mite_dat)
test_ids <- sample(1:n, 2)
train_x <- mite_dat[-test_ids, c("SubsDens", "WatrCont")]
train_y <- mite_dat$abundance[-test_ids]
test_x <- mite_dat[test_ids, c("SubsDens", "WatrCont")]
test_y <- mite_dat$abundance[test_ids]
head(train_x) SubsDens WatrCont
1 39.18 350.15
2 54.99 434.81
3 46.07 371.72
4 48.19 360.50
5 23.55 204.13
6 57.32 311.55Mite data: KNN results
- Running KNN with \(K = 3\) and using Euclidean distance, I identify the following neighbor sets for each test point:
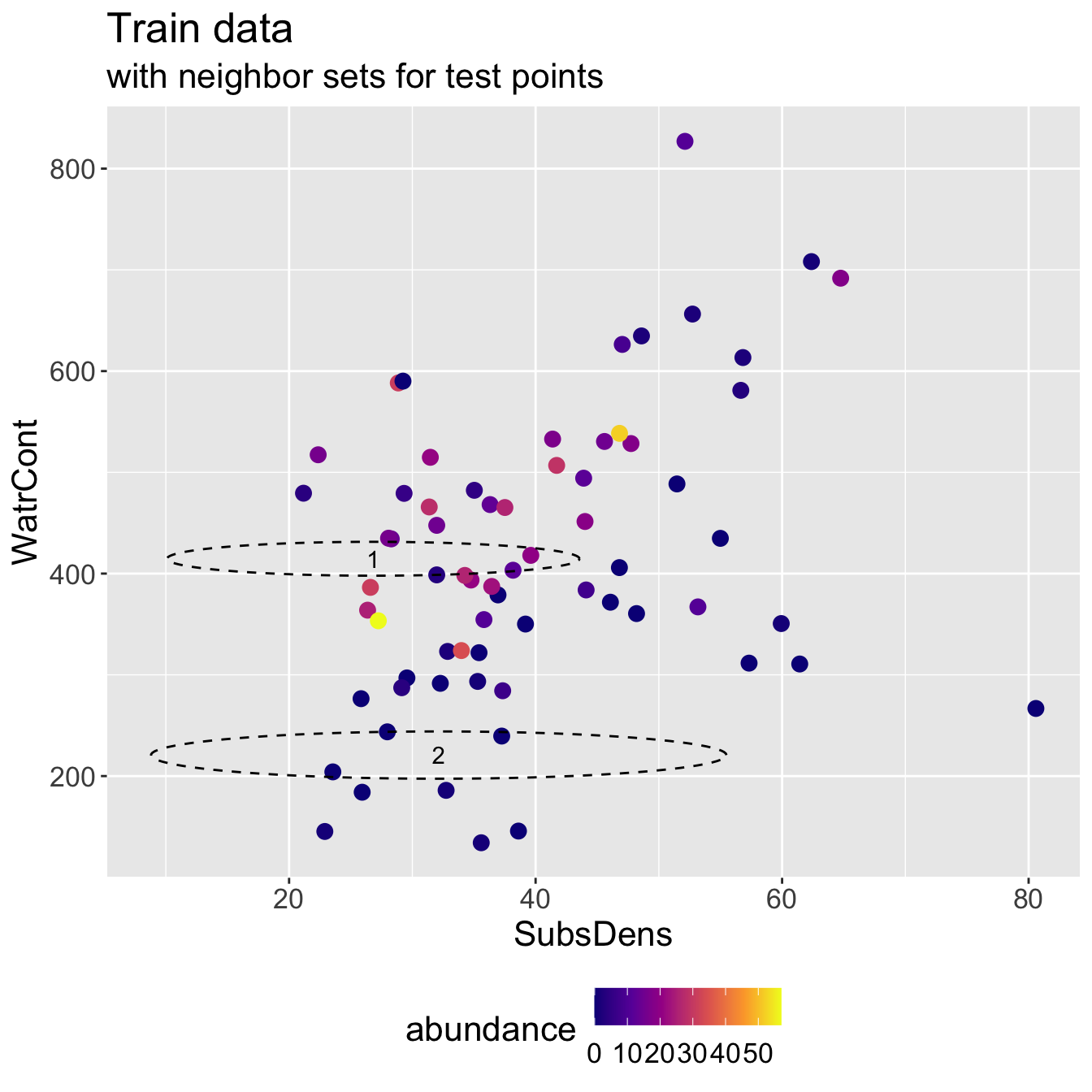
- Predicted abundance \(\hat{y}\) and true abundance \(y\) for both test points, for a test RMSE of 9.428.
# A tibble: 2 × 3
test_pt y_hat y_true
<int> <dbl> <int>
1 1 11.7 25
2 2 0 0Discuss: it seems like we did poorly for the first test observation. Does its neighbor set “make sense”?
Standardizing predictors
We will standardize our predictors, meaning that each predictor \(X_{j}\) will be transformed to have mean 0 and standard deviation 1:
\[X_{j}^{\text{std}} = \frac{X_{j} - \bar{X_{j}}}{\sigma_{X_{j}}},\]
where \(X_{j}\) is the vector of the \(j\)-th predictor, \(\bar{X}_{j}\) is the average of \(X_{j}\), and \(\sigma_{X_{j}}\) is its standard deviation.
[,1]
[1,] -0.032638509
[2,] 1.286001417
[3,] 0.542024937
[4,] 0.718844459
[5,] -1.336265457
[6,] 1.480336080
[7,] -0.218632629
[8,] 3.421180550
[9,] 1.823132417
[10,] -0.332064020
[11,] 0.602910905
[12,] -0.967613434
[13,] -0.193610999
[14,] 1.698024265
[15,] -0.347076999
[16,] -0.834998793
[17,] -1.145267011
[18,] 0.377716231
[19,] -0.080179607
[20,] -1.137760522
[21,] -0.569769510
[22,] -0.633157640
[23,] -0.608970064
[24,] -0.356251597
[25,] 0.992414286
[26,] -1.390478989
[27,] -0.313714825
[28,] -0.559760858
[29,] -0.116877998
[30,] 0.361035144
[31,] -0.186938564
[32,] -0.261169401
[33,] 1.134203525
[34,] -0.870029076
[35,] 0.003225828
[36,] -0.401290531
[37,] 0.681312014
[38,] 2.100038461
[39,] -0.442993249
[40,] -0.272012107
[41,] -1.081878880
[42,] 1.424454439
[43,] 1.902367580
[44,] 0.603744959
[45,] 0.370209741
[46,] -0.466346771
[47,] 0.178377241
[48,] -1.101062130
[49,] -0.940923695
[50,] -0.171925585
[51,] -0.893382597
[52,] 0.501990329
[53,] -0.633157640
[54,] 0.150853448
[55,] 1.438633362
[56,] -1.028499402
[57,] 1.097505134
[58,] -0.684034956
[59,] 0.622094155
[60,] 0.752206633
[61,] -0.378771064
[62,] -0.959272891
[63,] -1.534770392
[64,] -0.676528467
[65,] 1.046627818
[66,] -0.861688532
[67,] -0.854182043
[68,] -1.435517924
attr(,"scaled:center")
[1] 39.57132
attr(,"scaled:scale")
[1] 11.98963# confirming we have mean 0 and sd 1
scaled_SubsDens <- scale(train_x$SubsDens)
mean(scaled_SubsDens)[1] -4.865389e-16[1] 1Standardizing multiple variables
train_x_scaled <- train_x
train_x_scaled$SubsDens <- scale(train_x$SubsDens)
train_x_scaled$WatrCont <- scale(train_x$WatrCont)
head(train_x_scaled) SubsDens WatrCont
1 -0.03263851 -0.4434260
2 1.28600142 0.1503860
3 0.54202494 -0.2921323
4 0.71884446 -0.3708303
5 -1.33626546 -1.4676219
6 1.48033608 -0.7141694Standardizing the test data
We should use the same statistics from the training data to scale the test data
- i.e. to standardize the \(j\)-th predictor of the test data, we should use the mean and standard deviation of the \(j\)-th predictor from the training data
Discuss: why not scale the predictors first, and then split into train/test sets?
- Important:
- I do not scale the response variable
- I scale after splitting into train/test
Scaled mite data
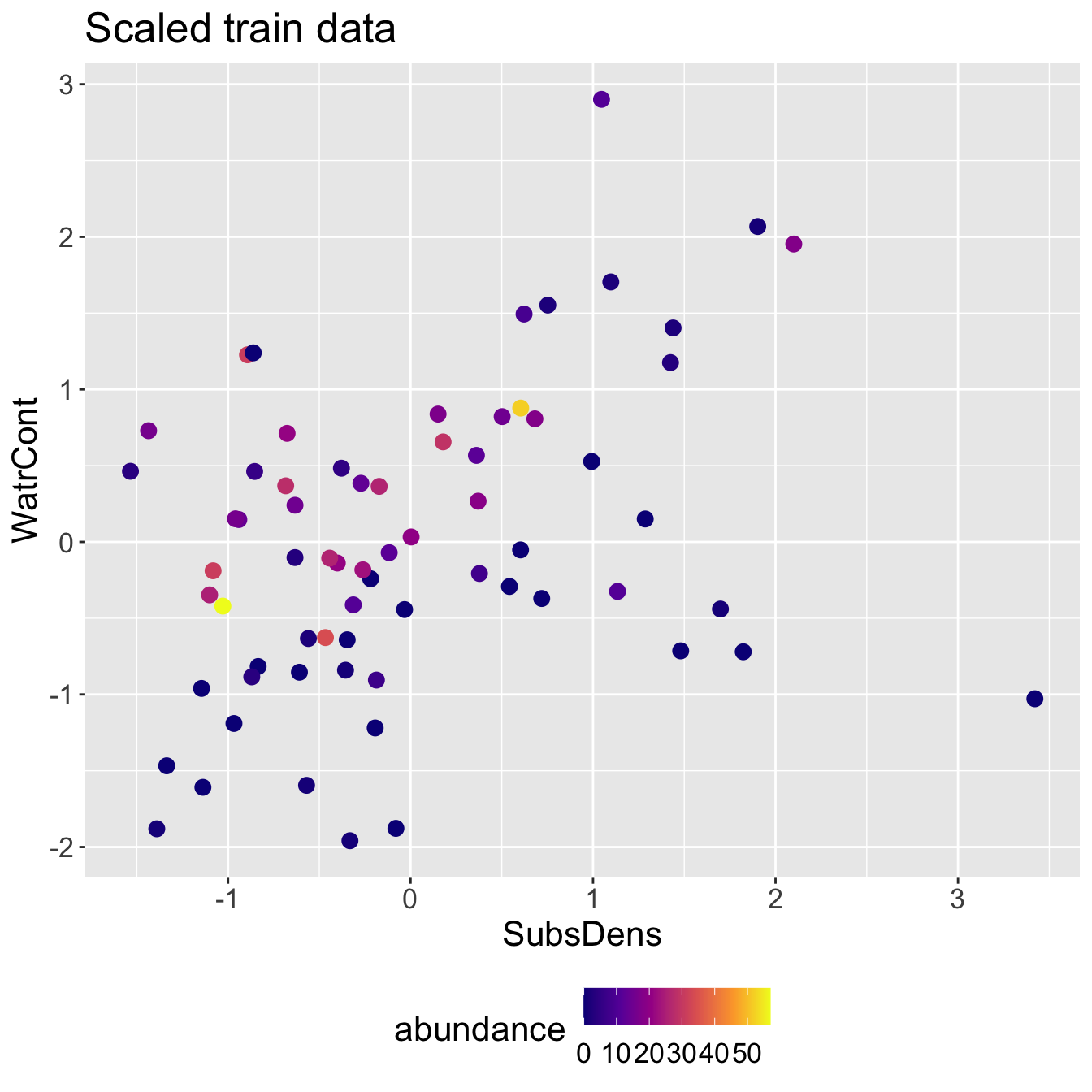
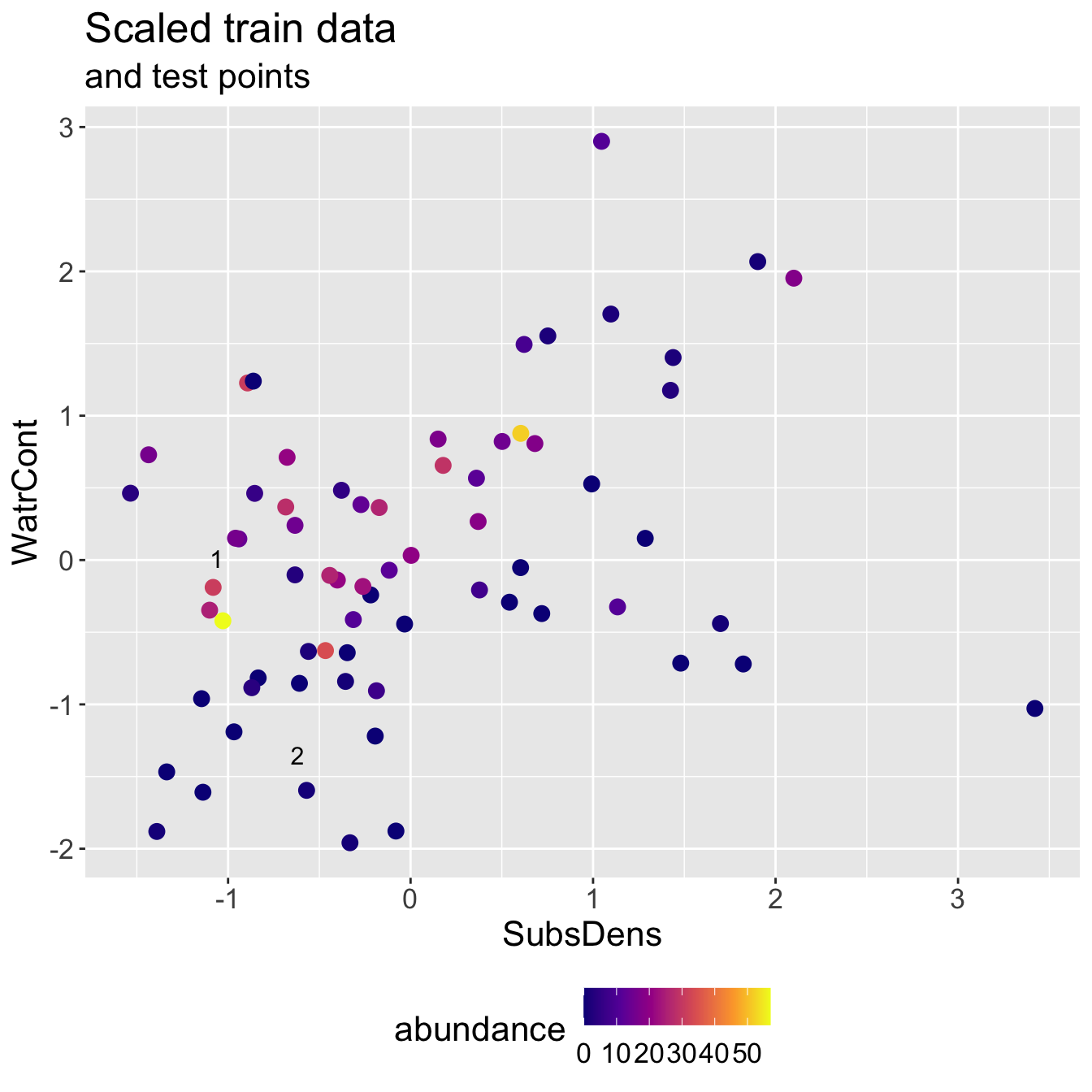
Scaled mite data: KNN results
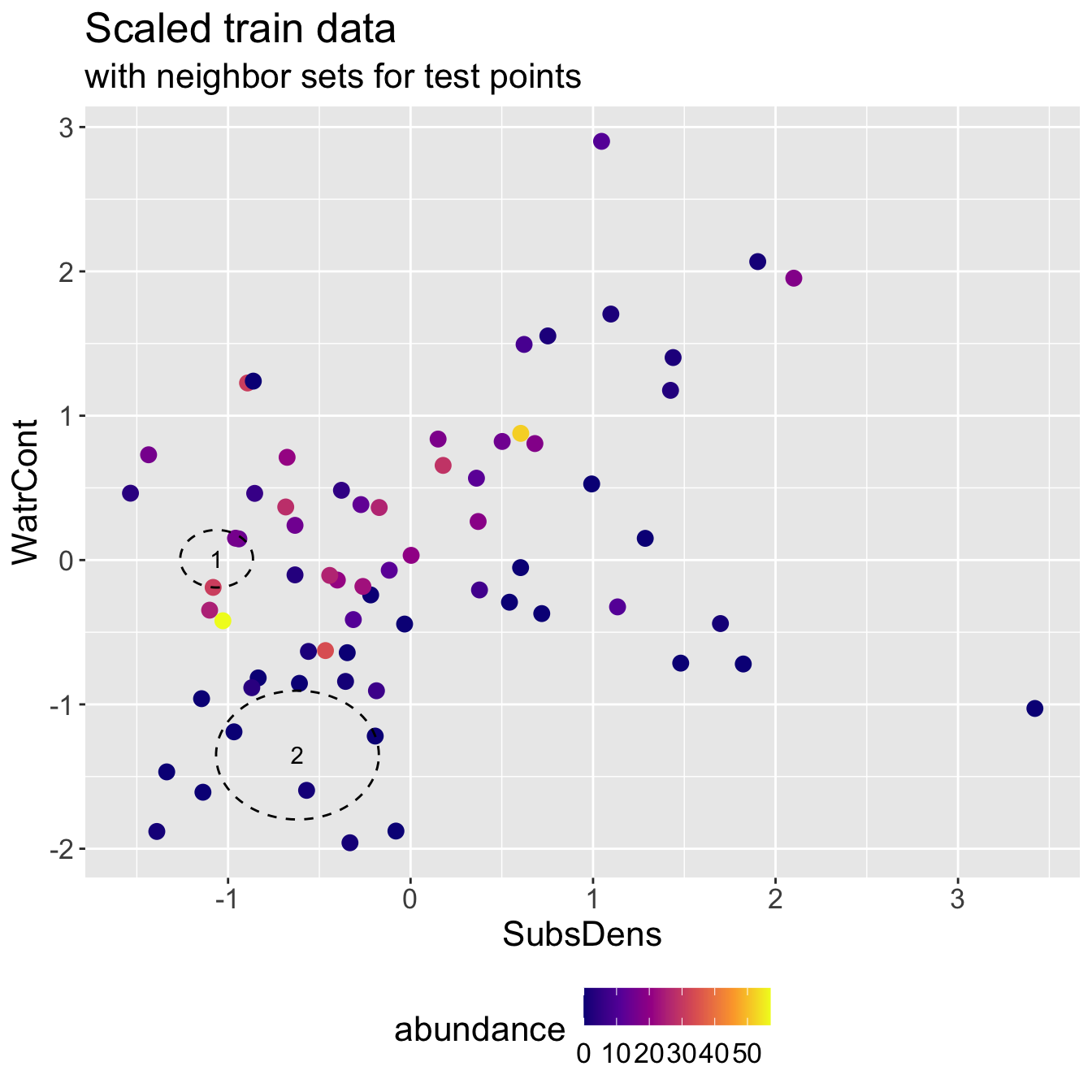
- Predicted abundance \(\hat{y}\) and true abundance \(y\) for both test points, for a test RMSE of 3.308.
# A tibble: 2 × 3
test_pt y_hat y_true
<int> <dbl> <int>
1 1 20.3 25
2 2 0.333 0Note how this RMSE compares to when we fit on original scale!
- Even though we do slightly worse predicting test point 2, we improve a lot on test point 1
Categorical predictors
Why are categorical predictors a problem?
Suppose we want to include the categorical predictors into our predictions:
Topo\(\in \{ \text{Blanket, Hummock}\}\)Shrub\(\in \{\text{None, Few, Many} \}\)Substrate\(\in \{\text{Sphagn1, Spaghn2, Sphagn3, Sphagn4, Litter, Barepeat, Interface}\}\)Discuss: how would you define “distance” or “closeness” between two observations based on:
TopoShrub
We will create new quantitative variables to represent categorical variables
Integer encoding
The predictor
Shrubis ordinal- i.e. there is a natural ordering (None < Few < Many) to the \(L = 3\) categories
The ordering should be reflected in the new quantitative variable
Integer encoding: convert each label (category/level) into an integer value, where:
- “Lowest” level gets assigned 0
- Second lowest level gets assigned 1
- …
- “Highest” level gets assigned \(L-1\)
Live code
Mite data: integer encoding
Shrub Shrub_encode
68 None 0
39 Few 1
1 Few 1
34 Many 2
43 Few 1- Now I can calculate distances using
Shrub_encode!
One-hot encoding
Question: why wouldn’t I want to use integer encoding for a non-ordinal variable such as
Substrate?One-hot encoding: map each level of the variable to a new binary 0/1 variable, where
- 0 represents the absence of the category
- 1 represents the presence of the category
These are called “dummy variables”; will have \(L\) new variables
Live code
Mite data: one-hot encoding
One-hot encoding of the Topo variable:
Topo Topo_hummock Topo_blanket
68 Blanket 0 1
39 Blanket 0 1
1 Hummock 1 0
34 Blanket 0 1
43 Blanket 0 1Mite data: one-hot encoding (cont.)
One-hot encoding of the Substrate variable:
# A tibble: 8 × 8
Substrate Sub_Sphagn1 Sub_Litter Sub_Interface Sub_S…¹ Sub_S…² Sub_S…³ Sub_B…⁴
<fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Sphagn2 0 0 0 0 0 1 0
2 Interface 0 0 1 0 0 0 0
3 Barepeat 0 0 0 0 0 0 1
4 Sphagn1 1 0 0 0 0 0 0
5 Sphagn1 1 0 0 0 0 0 0
6 Interface 0 0 1 0 0 0 0
7 Sphagn2 0 0 0 0 0 1 0
8 Interface 0 0 1 0 0 0 0
# … with abbreviated variable names ¹Sub_Sphagn3, ²Sub_Sphagn4, ³Sub_Sphagn2,
# ⁴Sub_BarepeatMite data: final data set
So, our final set of predictors X that we could use in KNN regression for the response variable abundance would be:
# A tibble: 70 × 12
SubsDens WatrCont Shrub_encode Topo_hummock Topo_blanket Sub_Sphagn1
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 46.8 406. 1 1 0 1
2 37.3 284. 2 0 1 0
3 29.2 590. 0 0 1 1
4 28.9 588. 0 0 1 1
5 46.8 539. 1 0 1 0
6 26.8 415. 0 0 1 0
7 47.0 626. 0 0 1 0
8 48.6 635. 0 0 1 0
9 56.6 581 1 0 1 0
10 27.2 353. 0 0 1 1
Sub_Litter Sub_Interface Sub_Sphagn3 Sub_Sphagn4 Sub_Sphagn2 Sub_Barepeat
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 0 0 0 0 0 0
2 0 1 0 0 0 0
3 0 0 0 0 0 0
4 0 0 0 0 0 0
5 0 1 0 0 0 0
6 0 1 0 0 0 0
7 0 1 0 0 0 0
8 0 1 0 0 0 0
9 0 1 0 0 0 0
10 0 0 0 0 0 0
# … with 60 more rows- Discuss: what are some potential issues with one-hot encoding?
Summary
Key point: when computing distances, you should consider standardizing your predictors if they are on very different scales
To calculate distances between categorical variables, we need to encode them somehow
Questions:
Should we standardize our new encoded variables?
Does anything need to change about your current KNN implementation to address standardizing variables and/or accommodating categorical predictors?