Linear Regression
2/23/23
Housekeeping
Lab 01 due tonight to Canvas at 11:59pm
Lab 02 assigned tomorrow (and has to do with baseball)
Linear regression
Linear regression
A simple, widely used approach in supervised learning
Assumes that the dependence of \(Y\) on the predictors \(X_{1}, \ldots, X_{p}\) is linear
Possible questions of interest
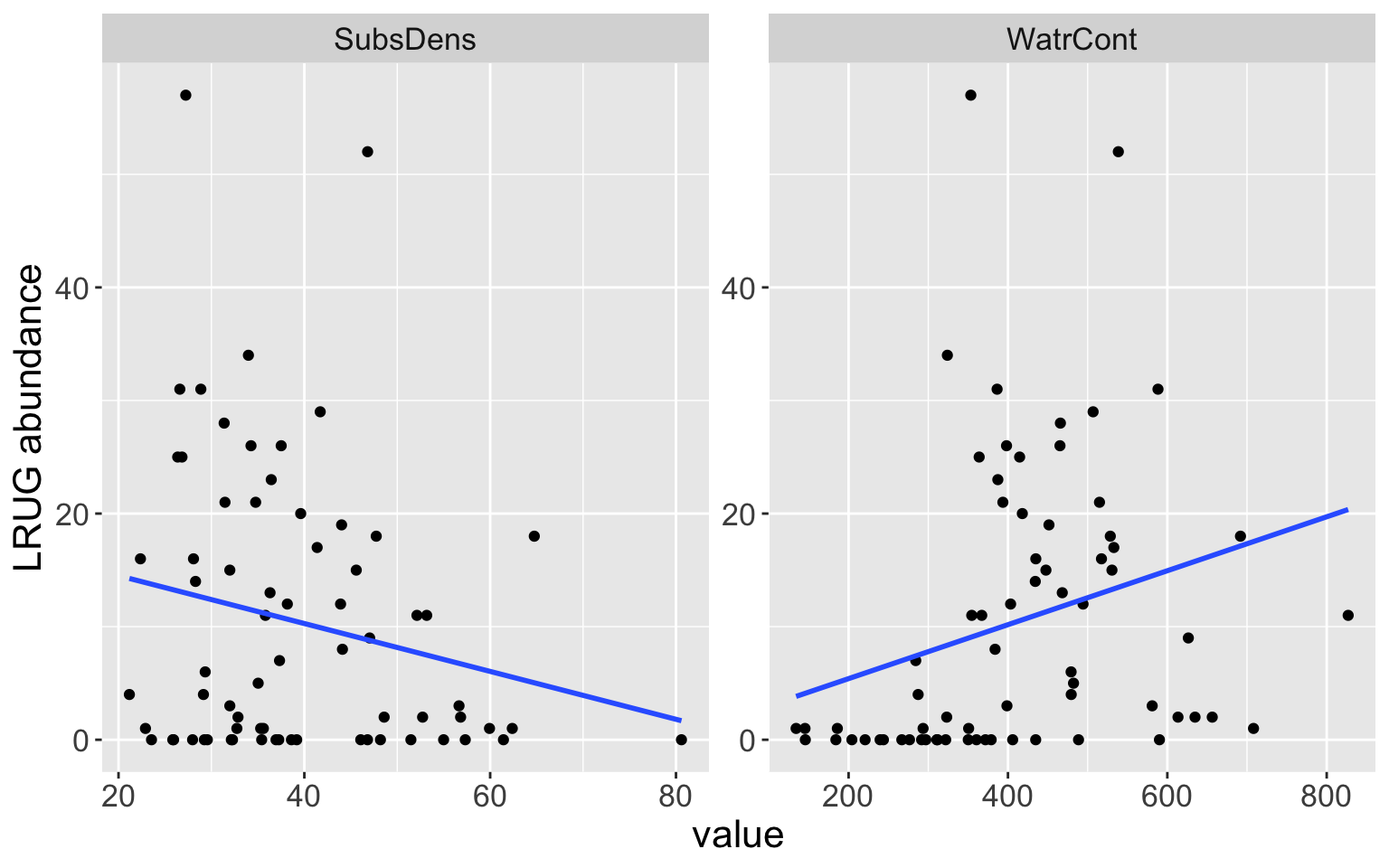
Is there a relationship between the abundance of LRUG mites and substrate density or water content in the soil where they are found?
- If so, how are strong are these relationships?
Is the relationship linear?
How accurately can we predict the abundance of these mites?
Simple Linear Regression
Simple linear regression
Simple linear regression (SLR)
Assumes a linear model for a quantitative response \(Y\) using a single predictor \(X\)
\[Y = \beta_{0} + \beta_{1} X + \epsilon,\]
where \(\beta_{0}, \beta_{1}\) are unknown coefficients (parameters) and \(\epsilon\) is the error
\(\beta_{0}\) is commonly referred to as the intercept, and \(\beta_{1}\) is the slope
For example:
abundance= \(\beta_{0}\) + \(\beta_{1}\)watercont+ \(\epsilon\)
Parameter estimation
Assuming \(n\) observations, we have data of the form \((x_{1}, y_{1}), (x_{2}, y_{2}), \ldots, (x_{n}, y_{n})\)
An SLR model says \[\begin{align*} y_{i} &= \beta_{0} + \beta_{1}x_{i}+ \epsilon\\ &\approx \beta_{0} + \beta_{1}x_{i}\ , \qquad \text{ for all } i = 1,\ldots, n \end{align*}\]
In this model, \(f(x_{i}) = \beta_{0} + \beta_{1} x_{i}\)
Notice that the relationship between \(x_{i}\) and \(y_{i}\) is the same for all \(i\)
In practice, \(\beta_{0}\) and \(\beta_{1}\) are unknown, so we must estimate them
Goal: obtain (good) estimates \(\hat{\beta}_{0}\) and \(\hat{\beta}_{1}\) that are as close to the true values as possible, such that \(y_{i} \approx \hat{\beta}_{0} + \hat{\beta}_{1} x_{i}\)
- How? Minimize the least squares criterion
Least squares
Let \(\hat{y}_{i} = \hat{\beta}_{0} + \hat{\beta}_{1}x_{i}\) be predicted response for \(i\)-th observation with predictor \(x_{i}\)
The \(i\)-th residual \(e_{i}\) is defined as \[e_{i} = y_{i} - \hat{y}_{i}\]
Define residual sum of squares (RSS) as \[\text{RSS} = e_{1}^{2} + e_{2}^{2} + \ldots + e_{n}^{2} = \sum_{i=1}^{n} e_{i}^2\]
Estimation by least squares
\[\text{RSS} = \sum_{i=1}^{n} e_{i}^2 = \sum_{i=1}^{n} (y_{i} - \hat{y}_{i})^2 = \sum_{i=1}^{n} (y_{i} - (\hat{\beta}_{0} + \hat{\beta}_{1}x_{i}))^2\]
Least squares approach selects the pair \((\hat{\beta}_{0}, \hat{\beta}_{1})\) that minimize the RSS. Can be shown that the minimizing values are: \[\begin{align*} \hat{\beta}_{1} &= \frac{\sum_{i=1}^{n}(x_{i} - \bar{x})(y_{i} - \bar{y})}{\sum_{i=1}^{n}(x_{i} - \bar{x})^2}\\ \hat{\beta}_{0} &= \bar{y} - \hat{\beta}_{1} \bar{x} \end{align*}\]
where \(\bar{y} = \frac{1}{n}\sum_{i=1}^{n}y_{i}\) and \(\bar{x} = \frac{1}{n}\sum_{i=1}^{n}x_{i}\)
Mite data
Least squares fit for abundance regressed on WaterCont, with residuals in orange.
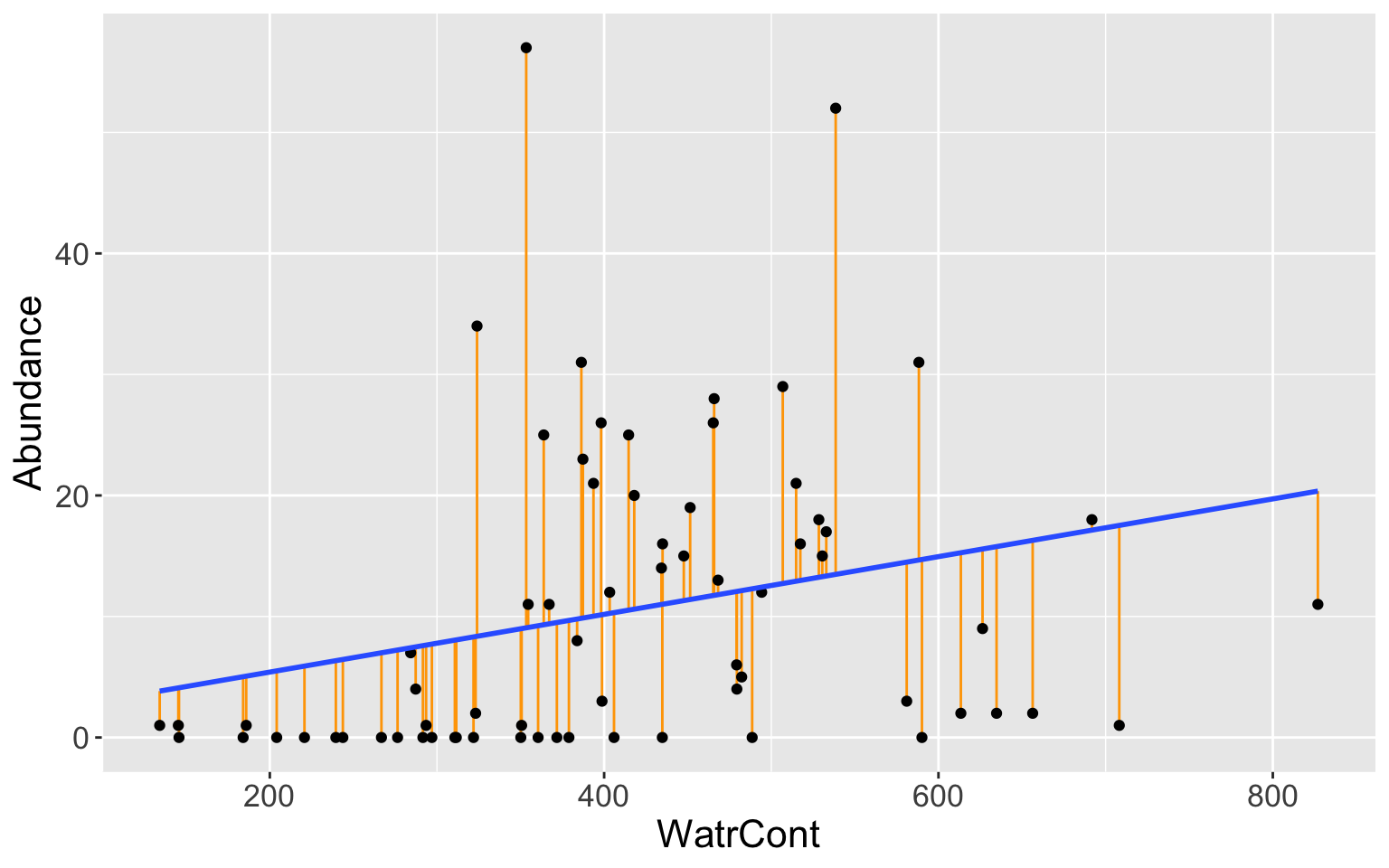
Let’s interpret this plot! Do you see anything strange or any patterns?
Interpreting Coefficients and Estimates
\[Y = \beta_{0} + \beta_{1} X + \epsilon\]
\(\beta_{0}\) is the expected value of \(Y\) when \(X = 0\)
\(\beta_{1}\) is the average increase in \(Y\) for one-unit increase in \(X\)
\(\epsilon\) is error
This equation is the population regression line
When using the least squares estimates for the coefficients, \(\hat{Y} = \hat{\beta}_{0} + \hat{\beta}_{1} X\) is the least squares line
Interpreting Coefficients and Estimates (cont.)
Call:
lm(formula = abundance ~ WatrCont, data = mite_dat)
Residuals:
Min 1Q Median 3Q Max
-16.525 -8.033 -4.088 4.493 47.937
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.63410 4.51171 0.141 0.8886
WatrCont 0.02385 0.01039 2.296 0.0248 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 12.29 on 68 degrees of freedom
Multiple R-squared: 0.07194, Adjusted R-squared: 0.05829
F-statistic: 5.271 on 1 and 68 DF, p-value: 0.02477\[\widehat{\text{LRUG}} = 0.634 + 0.024 \text{WatrCont}\]
How do I interpret \(\hat{\beta}_{0}\) and \(\hat{\beta}_{1}\) for this specific example?
Note: the estimates \(\hat{\beta}_{0}\) and \(\hat{\beta}_{1}\) will depend on the observed data! If I took a different sample of
LRUGmites, I would probably have different estimated values.- Isn’t that problematic? Can we asses how accurate are our estimates \(\hat{\beta}_{0}\) and \(\hat{\beta}_{1}\)?
Assessing Accuracy of Coefficient Estimates
Standard error (SE) of an estimator reflects how it varies under repeated sampling.
For simple linear regression: \[\text{SE}(\hat{\beta}_{0}) = \sigma^2 \left[ \frac{1}{n} + \frac{\bar{x}^2}{\sum_{i=1}^{n}(x_{i} - \bar{x})^2}\right] \qquad \text{SE}(\hat{\beta}_{1}) = \frac{\sigma^2}{\sum_{i=1}^{n}(x_{i} - \bar{x})^2}\]
where \(\sigma^2 = \text{Var}(\epsilon)\)
Typically \(\sigma^2\) is not known, but can be estimated from the data.
Estimate \(\hat{\sigma}\) is residual standard error (RSE), given by: \[\hat{\sigma}= \text{RSE} = \sqrt{\frac{1}{n-2}\text{RSS}}\]
We use this estimate to calculate \(\text{SE}(\hat{\beta}_{0})\) and \(\text{SE}(\hat{\beta}_{1})\)
Hypothesis Testing
Hypothesis testing is a method of statistical inference to determine whether the data at hand sufficiently support a particular hypothesis
Helps test the results of an experiment or survey to see if you have meaningful results
Helps draw conclusions about a population parameter
Standard errors can be used to perform hypothesis tests on the coefficients
Hypothesis Testing
Notion of “null” versus “alternate” hypothesis
Null hypothesis \(H_{0}\): there is no relationship between \(X\) and \(Y\)
Alternative hypothesis \(H_{A}\): there is some relationship between \(X\) and \(Y\)
Mathematically, corresponds to testing \[H_{0}: \beta_{1} = 0 \quad \text{ vs. } \quad H_{A}: \beta_{1} \neq 0\]
because if \(H_{0}\) true, then the model reduces to \(Y = \beta_{0} + \epsilon\) so there is no relationship
To test this null hypothesis, want to determine if \(\hat{\beta}_{1}\) is sufficiently far from zero
- How much is ‘sufficiently far’? Depends on \(\text{SE}(\hat{\beta}_{1})\).
p-value
With lots of hand-waving: can calculate a p-value, which is a probability that we observed the data we did, given that \(H_{0}\) is true. If the p-value is small, the observed data don’t seem to support \(H_{0}\)
Compare \(p\)-value to a pre-determined rejection level \(\alpha\) (often 0.05).
If \(p\)-value \(< \alpha\), reject \(H_{0}\). Otherwise, fail to reject \(H_{0}\).
Call:
lm(formula = abundance ~ WatrCont, data = mite_dat)
Residuals:
Min 1Q Median 3Q Max
-16.525 -8.033 -4.088 4.493 47.937
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.63410 4.51171 0.141 0.8886
WatrCont 0.02385 0.01039 2.296 0.0248 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 12.29 on 68 degrees of freedom
Multiple R-squared: 0.07194, Adjusted R-squared: 0.05829
F-statistic: 5.271 on 1 and 68 DF, p-value: 0.02477\(H_{0}: \beta_{1} = 0\) (there is no relationship between
LRUGabundance andWatrCont)- What is the p-value for this hypothesis?
- What is our decision in the mite example?
Why hypothesis testing?
Hypothesis testing can help us determine if there is a relationship between the predictor and the response variable!
This is the inference part of statistical learning
If there is a relationship, then it makes sense to interpret the strength of the relationship (i.e. interpret the value \(\hat{\beta}_{1}\))
Multiple Linear Regression
Multiple linear regression
In practice, we often have more than one predictor
With \(p\) predictors, the model is \[Y = \beta_{0} + \beta_{1}X_{1} + \beta_{2}X_{2} + \ldots + \beta_{p}X_{p} +\epsilon\]
Interpret \(\beta_{j}\) as the average effect on \(Y\) for a one-unit increase in \(X_{j}\), holding all other predictors fixed/constant
Mite data
Regressing abundance on both WatrCont and SubsDens:
Call:
lm(formula = abundance ~ WatrCont + SubsDens, data = mite_dat)
Residuals:
Min 1Q Median 3Q Max
-20.192 -8.633 -1.385 6.866 44.245
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 10.30549 5.48833 1.878 0.06477 .
WatrCont 0.03444 0.01057 3.257 0.00177 **
SubsDens -0.35682 0.12604 -2.831 0.00612 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 11.7 on 67 degrees of freedom
Multiple R-squared: 0.1711, Adjusted R-squared: 0.1464
F-statistic: 6.915 on 2 and 67 DF, p-value: 0.001861\[\widehat{abundance} = 10.306 + 0.034 \text{WatrCont} -0.357 \text{SubsDens}\]
How do we interpret the estimated coefficients?
How do we interpret the p-values?
- The p-value for \(\beta_{j}\) corresponds to the test of \(H_{0}: \beta_{j} = 0\) and \(\beta_{k} (k \neq j)\) unrestricted
Model assessment
Model fit
How well does our linear regression model “fit” the data it was trained on? How accurate is our model?
Residual standard error (RSE) \[\text{RSE} = \sqrt{\frac{1}{n-p-1} \sum_{i=1}^{n} (y_{i} - \hat{y}_{i})^2},\]
where \(p\) is the number of predictors, and \(i\) indexes the observations used to fit the model
RSE is considered a measure of the lack of fit of the model
- Measured in the units of \(Y\)
Predictions
How well does our linear regression model predict new responses for a given set of covariates?
For example, suppose I want to use our model to predict the abundance of
LRUGmites at a new sampling location where theWatrContis 400 g/L and theSubsDensis 30 g/LI will plug these values into our fitted model
m2:
\[\widehat{abundance} = 10.306 + 0.034 \times 400 -0.357 \times 30 = 13.3747829\]
- Okay…great! But should I trust these predictions?
Prediction performance
We could get a better sense of a model’s prediction performance by comparing the predicted responses to the true values
We should always compare prediction performance for “previously unseen” data (i.e. test data)
- The model already “knows” the data used to fit it (i.e. the training data)
Discuss: what are some important criteria for the testing data?
Qualitative predictors
Qualitative predictors
Thus far, we have assumed that all predictors in our linear model are quantitative. In practice, we often have categorical predictors
Our mite data has the following categorical variables:
Shrub,Substrate, andTopoLet’s begin with the simplest case: a categorical predictor with two categories/levels
- e.g. the
Topovariable takes on the values “Blanket” or “Hummock” only
- e.g. the
2-level qualitative predictor
We will create an indicator or dummy variable as follows: \[\text{TopoBlanket}_{i} = \begin{cases} 1 & \text{ if } \color{blue}{\text{Topo}_{i}} = \text{Blanket} \\ 0 & \text{ if } \color{blue}{\text{Topo}_{i}} = \text{Hummock} \end{cases}\]
Simple linear regression model for
LRUGregressed onTopo: \[\text{LRUG}_{i} = \beta_{0} + \beta_{1}\text{TopoBlanket}_{i} + \epsilon_{i} = \begin{cases} \beta_{0} + \epsilon_{i} & \text{ if } \color{blue}{\text{Topo}_{i}} = \text{Blanket} \\ \beta_{0} + \beta_{1} + \epsilon_{i} & \text{ if } \color{blue}{\text{Topo}_{i}} = \text{Hummock} \end{cases}\]How to interpret?
Mite data
Call:
lm(formula = abundance ~ Topo, data = mite_dat)
Residuals:
Min 1Q Median 3Q Max
-15.318 -4.318 -2.154 4.473 41.682
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 15.318 1.658 9.238 1.26e-13 ***
TopoHummock -13.164 2.721 -4.838 7.85e-06 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 11 on 68 degrees of freedom
Multiple R-squared: 0.2561, Adjusted R-squared: 0.2452
F-statistic: 23.41 on 1 and 68 DF, p-value: 7.851e-06Fitted model is: \[\widehat{\text{LRUG}} = 15.318 - 13.164 \text{TopoHummock}\]
Interpret!
More than two levels
With more than two levels, we simply create additional dummy variables: \[\begin{align*}\text{Shrub}_{i,Few} &= \begin{cases} 1 & \text{ if } \color{blue}{\text{Shrub}_{i}} = \text{Few} \\ 0 & \text{ if } \color{blue}{\text{Shrub}_{i}} = \text{not Few} \end{cases} \\ \text{Shrub}_{i, Many} &= \begin{cases} 1 & \text{ if } \color{blue}{\text{Shrub}_{i}} = \text{Many} \\ 0 & \text{ if } \color{blue}{\text{Shrub}_{i}} = \text{not Many} \end{cases} \end{align*}\]
Resulting regression model for
LRUGwith onlyShrubas predictor: \[\begin{align*} \text{LRUG}_{i} &= \beta_{0} + \beta_{1} \text{Shrub}_{i, Few} + \beta_{2} \text{Shrub}_{i, Many} + \epsilon_{i}\\ &\approx \begin{cases} \beta_{0} + \beta_{1} & \text{ if } \color{blue}{\text{Shrub}_{i}} = \text{Few} \\ \beta_{0} + \beta_{2} & \text{ if } \color{blue}{\text{Shrub}_{i}} = \text{Many} \\ \beta_{0} & \text{ if } \color{blue}{\text{Shrub}_{i}} = \text{None} \end{cases}\end{align*}\]
Call:
lm(formula = abundance ~ Shrub, data = mite_dat)
Residuals:
Min 1Q Median 3Q Max
-16.895 -8.060 -2.760 6.635 40.105
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 10.923 1.405 7.775 6.12e-11 ***
Shrub.L -9.288 2.505 -3.707 0.000427 ***
Shrub.Q -1.460 2.359 -0.619 0.538164
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 11.64 on 67 degrees of freedom
Multiple R-squared: 0.1791, Adjusted R-squared: 0.1545
F-statistic: 7.306 on 2 and 67 DF, p-value: 0.001348Remarks
For a given categorical variable, there will always be one fewer dummy variables than levels
- Level with no dummy variable is known as baseline. For model
m4on previous slide, the “None” category was the baseline level
- Level with no dummy variable is known as baseline. For model
Can have multiple categorical variables in a single model:
Call:
lm(formula = abundance ~ Topo + Shrub, data = mite_dat)
Residuals:
Min 1Q Median 3Q Max
-16.895 -6.056 0.691 4.756 40.105
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 14.755 1.641 8.993 4.48e-13 ***
TopoHummock -11.253 2.997 -3.755 0.000369 ***
Shrub.L -4.832 2.580 -1.873 0.065552 .
Shrub.Q -3.128 2.203 -1.420 0.160385
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 10.65 on 66 degrees of freedom
Multiple R-squared: 0.3236, Adjusted R-squared: 0.2928
F-statistic: 10.52 on 3 and 66 DF, p-value: 9.593e-06Let’s write out the estimated regression model together
What does \(\beta_{0}\) represent?
How do we interpret the coefficients?
Extensions of linear model (optional)
Linear model is restrictive
Linear model is widely used and works quite well, but has several highly restrictive assumptions
- Relationship between \(X\) and \(Y\) is additive
- Relationship between \(X\) and \(Y\) is linear
There are common approaches to loosen these assumptions. We will only discuss the first restriction here. Take a regression class for more!
Interactions
- Additive assumption: the association between a predictor \(X_{j}\) and the response \(Y\) does not depend on the value of any other predictors
\[Y = \beta_{0} + \beta_{1} X_{1} + \beta_{2} X_{2} + \epsilon\]
vs
\[Y = \beta_{0} + \beta_{1} X_{1} + \beta_{2} X_{2} + \beta_{3}\color{orange}{X_{1}X_{2}} + \epsilon\]
This third predictor \(\color{orange}{X_{1}X_{2}}\) is known as an interaction term
The total effect of \(X_{1}\) on \(Y\) also depends on the value of \(X_{2}\) through the interaction
In the above equation, \(\beta_{1}\) and \(\beta_{2}\) are called the “main effects” of \(X_{1}\) and \(X_{2}\) respectively
Mite data
- Suppose I want to regress
LRUGabundance onSubsDensandWatrContand their interaction:
Call:
lm(formula = abundance ~ SubsDens + WatrCont + SubsDens * WatrCont,
data = mite_dat)
Residuals:
Min 1Q Median 3Q Max
-24.012 -7.155 -1.896 5.011 44.539
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -1.012e+01 1.369e+01 -0.740 0.4621
SubsDens 1.323e-01 3.256e-01 0.406 0.6859
WatrCont 8.667e-02 3.379e-02 2.565 0.0126 *
SubsDens:WatrCont -1.210e-03 7.443e-04 -1.625 0.1088
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 11.56 on 66 degrees of freedom
Multiple R-squared: 0.203, Adjusted R-squared: 0.1668
F-statistic: 5.604 on 3 and 66 DF, p-value: 0.001744Mite data
Fitted model: \(\widehat{LRUG} = -10.12 + 0.132 \text{SubsDens} + 0.087 \text{WatrCont} -0.001 \text{SubsDens} \times \text{WatrCont}\)
Interpretations?
- Estimates suggest that a 1 g/L increase in the \(\color{blue}{\text{WaterCont}}\) of the soil is associated with an increased abundance of (0.087 + -0.001 \(\times \color{blue}{\text{SubsDens}}\)) LRUG mites
Interactions with categorical variable
Can also have interactions involving categorical variables!
In particular, the interaction between a quantitative and a categorical variable has nice interpretation
Consider the effects of
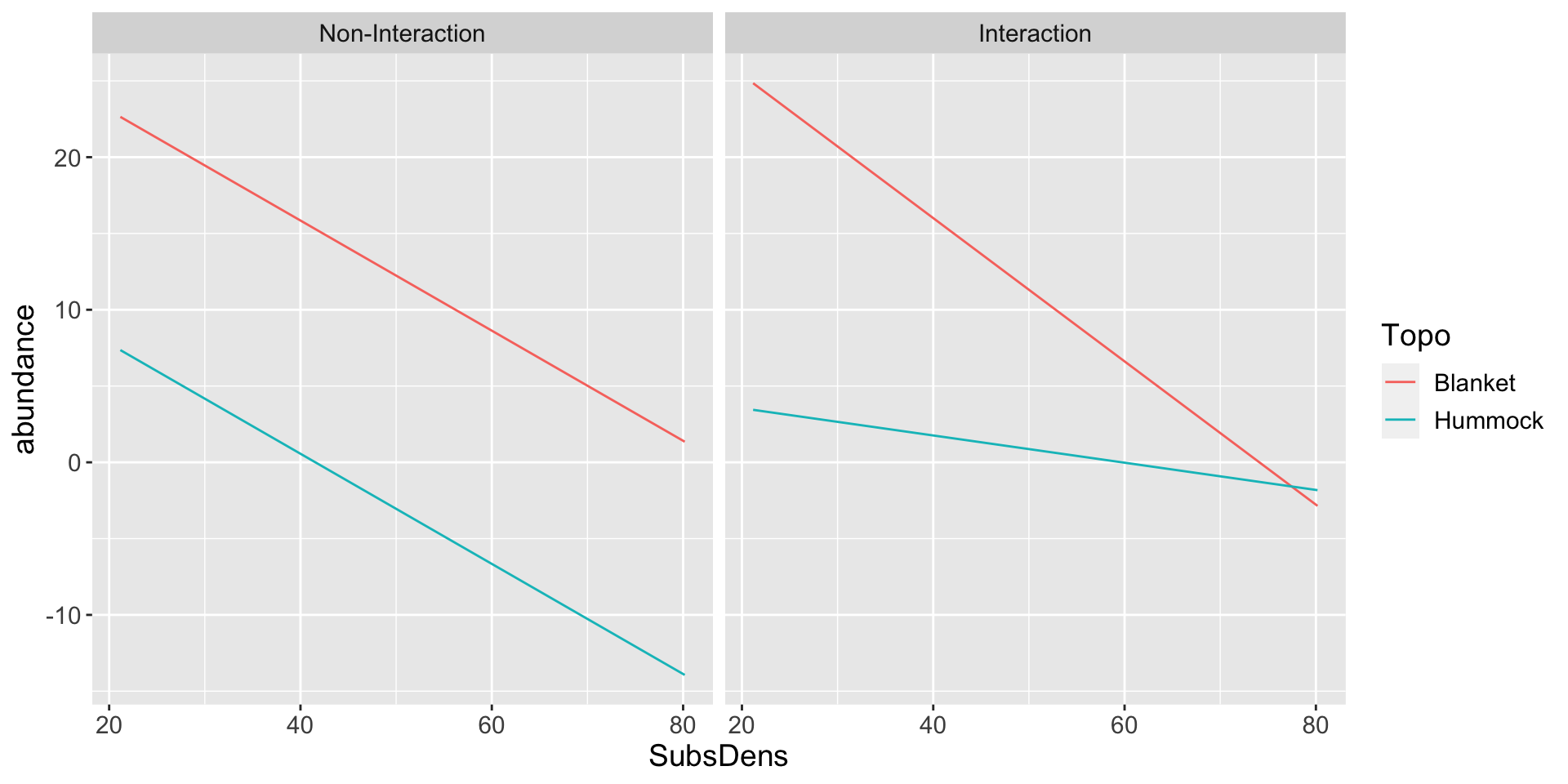
SubsDensandTopoand their interaction on the abundances: \[\begin{align*} \text{LRUG}_{i} &\approx \beta_{0} + \beta_{1} \text{SubsDens}_{i} + \beta_{2}\text{TopoBlanket}_{i} + \beta_{3} \text{SubsDens}_{i} \times \text{TopoBlanket}_{i} \\ & = \begin{cases} \beta_{0} + \beta_{1}\text{SubsDens}_{i} & \text{ if } \color{blue}{\text{Topo}_{i}} = \text{Blanket} \\ (\beta_{0} + \beta_{2}) + (\beta_{1} + \beta_{3}) \text{SubsDens}_{i} & \text{ if } \color{blue}{\text{Topo}_{i}} = \text{Hummock} \end{cases} \end{align*}\]
Call:
lm(formula = abundance ~ SubsDens + Topo + SubsDens * Topo, data = mite_dat)
Residuals:
Min 1Q Median 3Q Max
-21.059 -4.764 -1.186 3.229 39.191
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 34.7881 5.3697 6.479 1.35e-08 ***
SubsDens -0.4695 0.1242 -3.781 0.000338 ***
TopoHummock -29.4584 9.0331 -3.261 0.001757 **
SubsDens:TopoHummock 0.3803 0.2323 1.637 0.106406
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 10.11 on 66 degrees of freedom
Multiple R-squared: 0.3901, Adjusted R-squared: 0.3624
F-statistic: 14.07 on 3 and 66 DF, p-value: 3.425e-07Fitted regression lines