Live code:
hclust()
We use the function hclust() to implement hierarchical clustering! We need to pass in two arguments:
A distance matrix of the dissimilarities between observations. The distance matrix must always be \(n \times n\), where \(n\) is the number of observations. The pairwise Euclidean distances are easily calculated used
dist().We also need to specify the type of linkage, in quotes
- “average”, “single”, “complete”, or “centroid” (there are others, but we didn’t discuss in class)
We will visit the USArrests data we saw in class. I will grab two variables, and go ahead and standardize them.
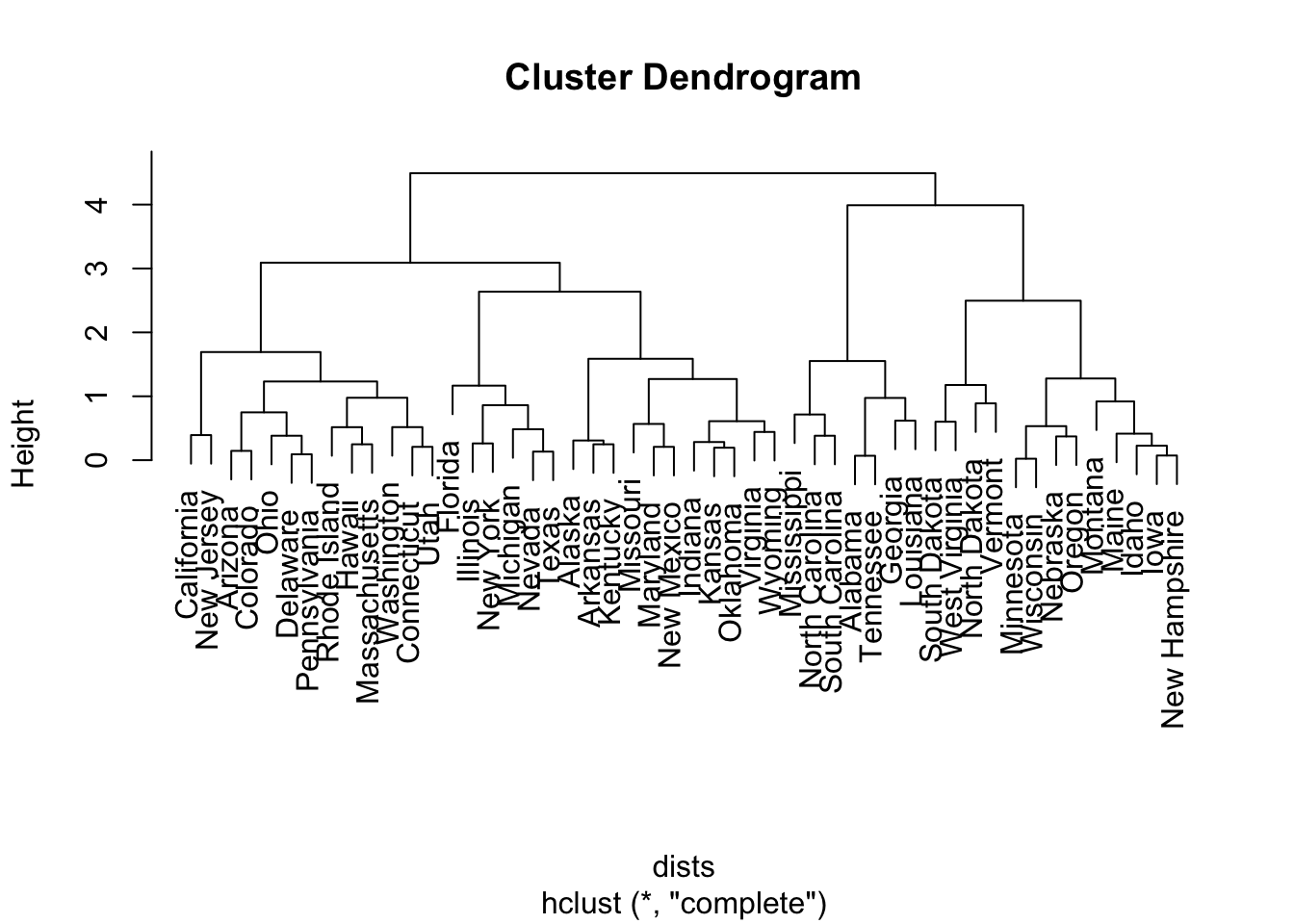
We can easily pass the output into the plot() function to visualize the entire dendrogram:
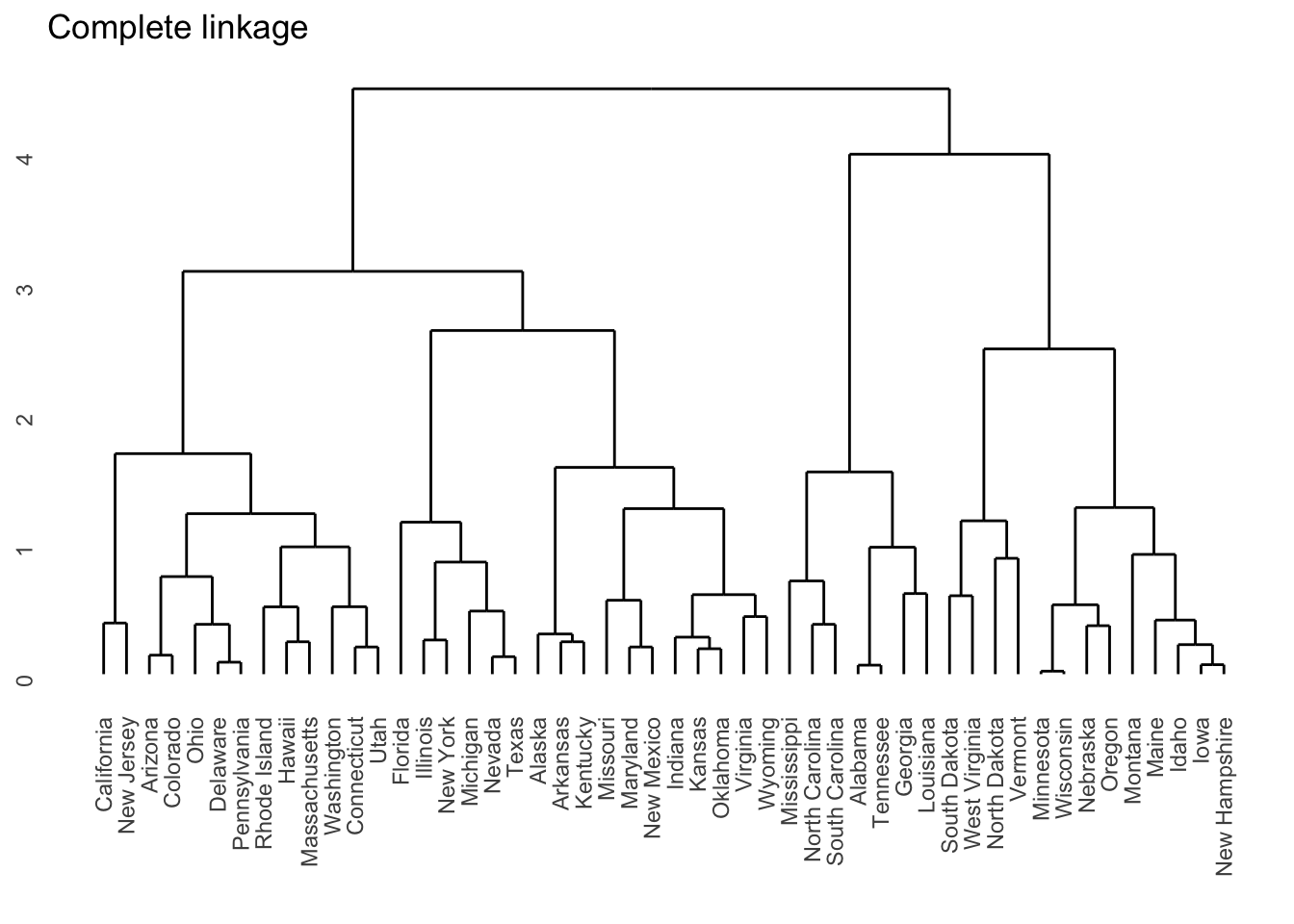
I personally prefer ggplots, so if you install the ggdendro package, you can obtain the following plot:
To determine the cluster labels associated with a given cut of the dendrogram, we use the cutree() function. It takes in the output from hclust() and the number of clusters k we want to obtain. In the following, I obtain three clusters:
Alabama Alaska Arizona Arkansas California
1 2 2 2 2
Colorado Connecticut Delaware Florida Georgia
2 2 2 2 1
Hawaii Idaho Illinois Indiana Iowa
2 3 2 2 3
Kansas Kentucky Louisiana Maine Maryland
2 2 1 3 2
Massachusetts Michigan Minnesota Mississippi Missouri
2 2 3 1 2
Montana Nebraska Nevada New Hampshire New Jersey
3 3 2 3 2
New Mexico New York North Carolina North Dakota Ohio
2 2 1 3 2
Oklahoma Oregon Pennsylvania Rhode Island South Carolina
2 3 2 2 1
South Dakota Tennessee Texas Utah Vermont
3 1 2 2 3
Virginia Washington West Virginia Wisconsin Wyoming
2 2 3 3 2 From the output, we can see to which of the 4 clusters each state has been assigned. To get an idea of the spread across the clusters, we can use the table() function:
Comparing to k-means
What happens if I run the k-means algorithm on the same set of data? Do we think the two methods will agree with each other if I specify I want the same number of clusters?
Now, let’s be careful! There’s no reason why cluster 1 from hierarchical clustering should be the “same” as cluster 1 from k-means. So if we want to compare the results from the two methods, we just need to see if a cluster from the hierarchical method has a lot of the same observations as a cluster from k-means.
Linkage choices
Try playing around with different choices of linkage! What do you notice if you use single linkage?