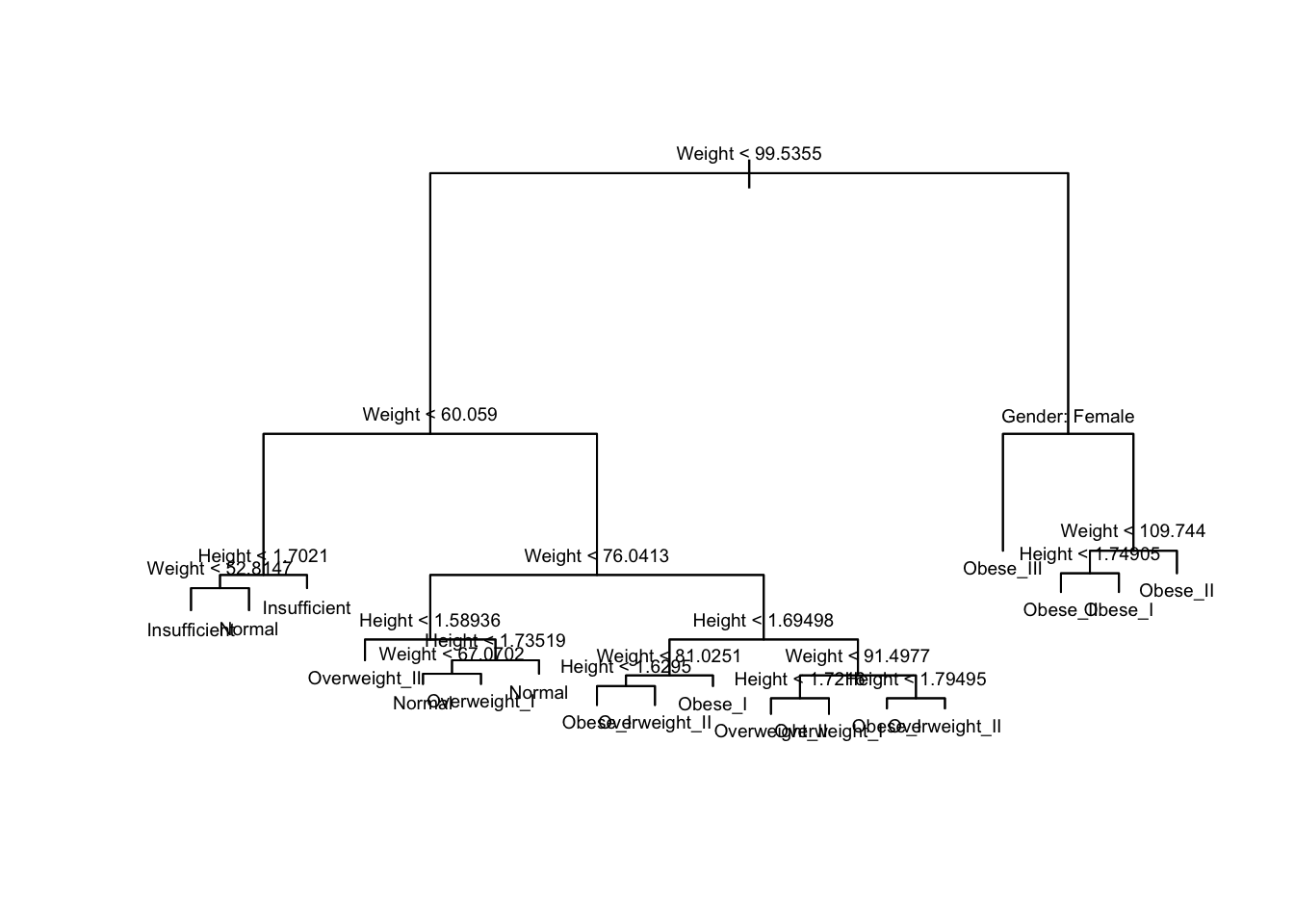
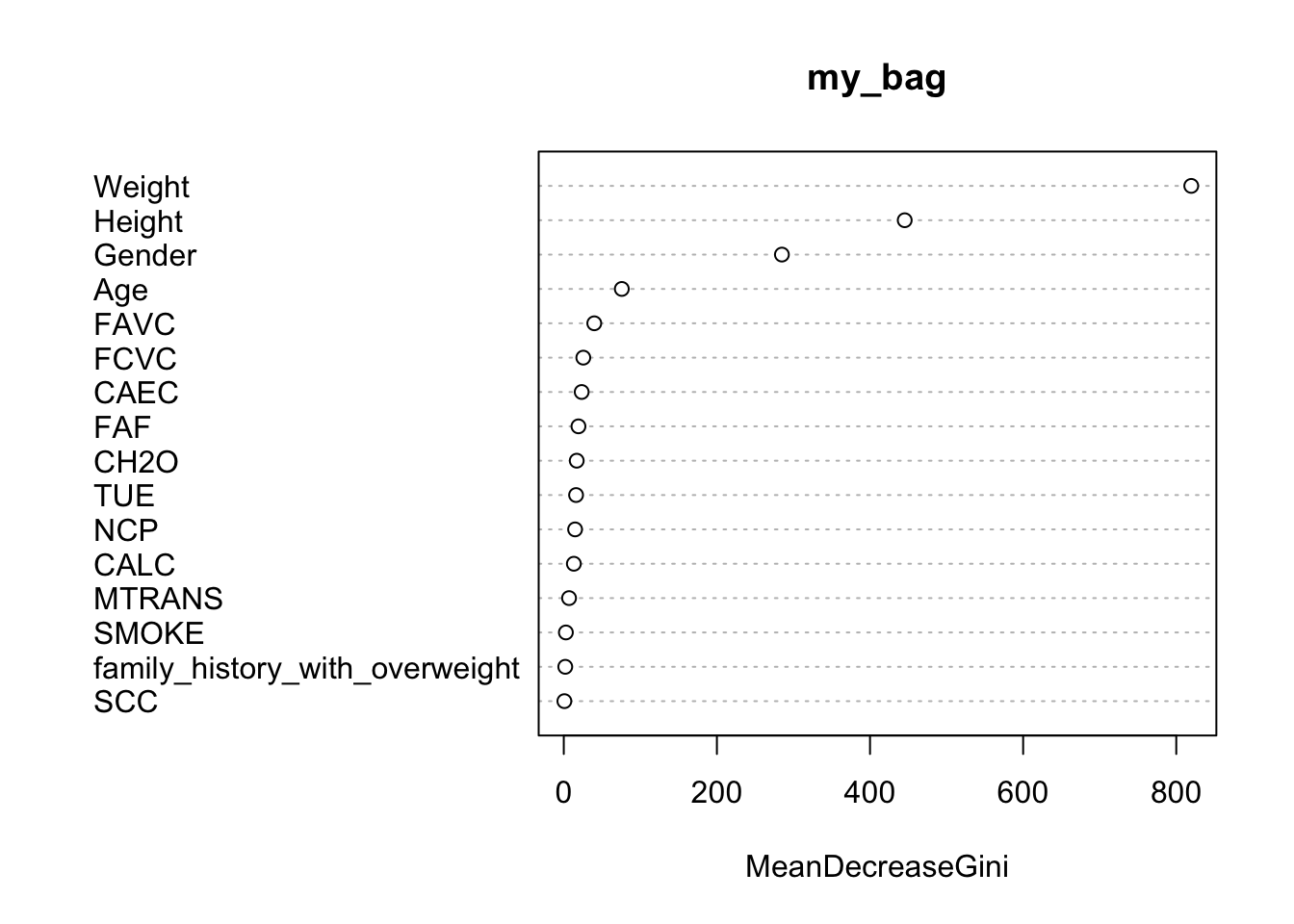
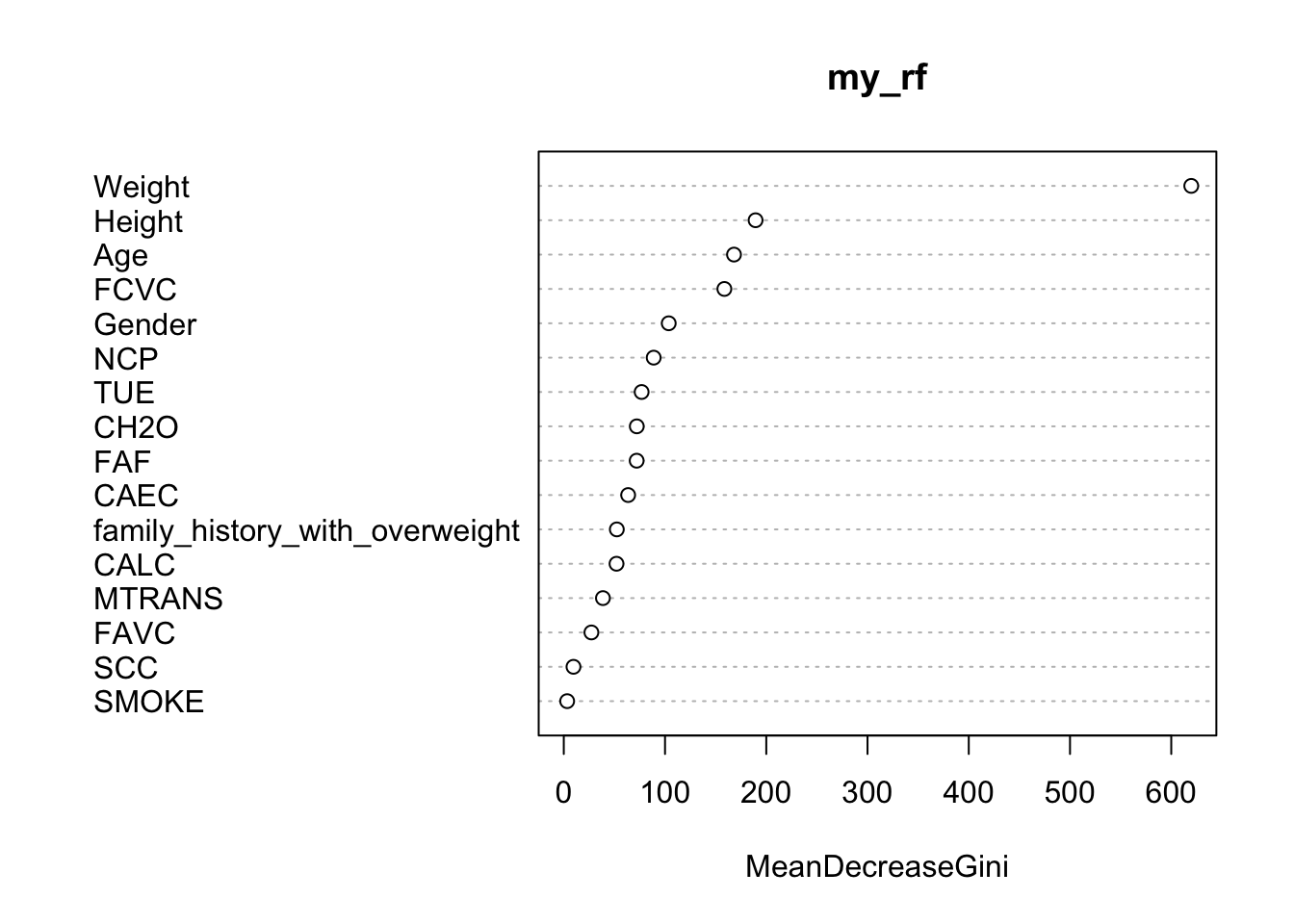
library(tidyverse)
obesity <- read.csv("https://raw.githubusercontent.com/math218-spring2023/class-data/main/obesity.csv")
obesity <- obesity %>%
mutate_if(is.character, factor)
obesity %>%
count(class) class n
1 Insufficient 272
2 Normal 287
3 Obese_I 351
4 Obese_II 297
5 Obese_III 324
6 Overweight_I 290
7 Overweight_II 290