Live code:
Introduction
We will use the R package called randomForest is fit bagged decision trees and random forests. Go ahead and install this package in your console!
As we will see in the slides, bagged trees and random forests are very similar and can be fit using the same function: randomForest(). The difference lies in the specification of the mtry argument, as we will see below.
Bagging
The syntax for bagged regression trees is the same as in regular regression trees: response ~ predictors.
In bagged regression trees, the only parameter that the modeler needs to choose is the number of bootstrap samples \(B\) to obtain (and therefore the number of trees to fit). This is denoted as the ntree argument
However, we will need to specify the additional mtry argument to specify we want to fit a bagged model rather than a random forest. For bagged trees, we set mtry equal to the number of predictors we have.
In the following code, I fit B = 10 regression trees, and specify mtry = 5.
To make predictions for the test set, we will use the familiar predict() function:
10 14 20 21 24 25 26 29
0.400000 3.675000 0.700000 5.916667 0.400000 4.125000 6.050000 4.166667
31 37 46 52 63 66
9.240000 24.790000 21.910000 16.800000 19.616667 23.231667 Out-of-bag error
The nice thing about bootstrapping is that typically ~1/3 of observations are left out in each sample (and therefore, in each one of the B trees). So, we don’t necessarily need to explicitly specify a test/train split!
In the following code, a fit a bagged model using all of the available observations:
The randomForest() function will automatically create a vector of predicted values for the input data based on the out of bag (OOB) samples; i.e. whenever observation \(i\) is OOB (not included in the bootstrap sample) for tree \(b\), we can treat \(i\) as a test observation and obtain a prediction for it. This is accessed through the predicted component of the fitted model:
1 2 3 4 5 6
3.86666667 5.27777778 8.30000000 3.75833333 0.00000000 0.31111111
7 8 9 10 11 12
3.61944444 2.79000000 0.60000000 0.15000000 2.00000000 0.88888889
13 14 15 16 17 18
0.00000000 5.92500000 33.70000000 2.06250000 1.52777778 0.66666667
19 20 21 22 23 24
32.80000000 1.37500000 4.56250000 0.08571429 12.01111111 2.31250000
25 26 27 28 29 30
0.40000000 7.05555556 0.65000000 7.80000000 0.30000000 19.75000000
31 32 33 34 35 36
4.58333333 0.13333333 21.30000000 16.61000000 NA 9.14285714
37 38 39 40 41 42
14.99000000 11.75000000 1.00000000 21.10000000 24.82000000 22.60000000
43 44 45 46 47 48
9.70833333 5.86666667 13.41666667 14.50000000 1.40000000 11.31000000
49 50 51 52 53 54
44.11666667 13.25000000 16.25000000 16.81250000 18.13333333 17.58333333
55 56 57 58 59 60
19.22000000 16.25000000 2.03666667 11.86666667 7.67000000 20.80000000
61 62 63 64 65 66
19.06666667 13.40714286 15.03333333 19.32857143 14.75000000 15.45000000
67 68 69 70
6.20000000 34.00000000 4.54666667 3.50000000 Do you notice anything strange in these predictions?
Importance measure
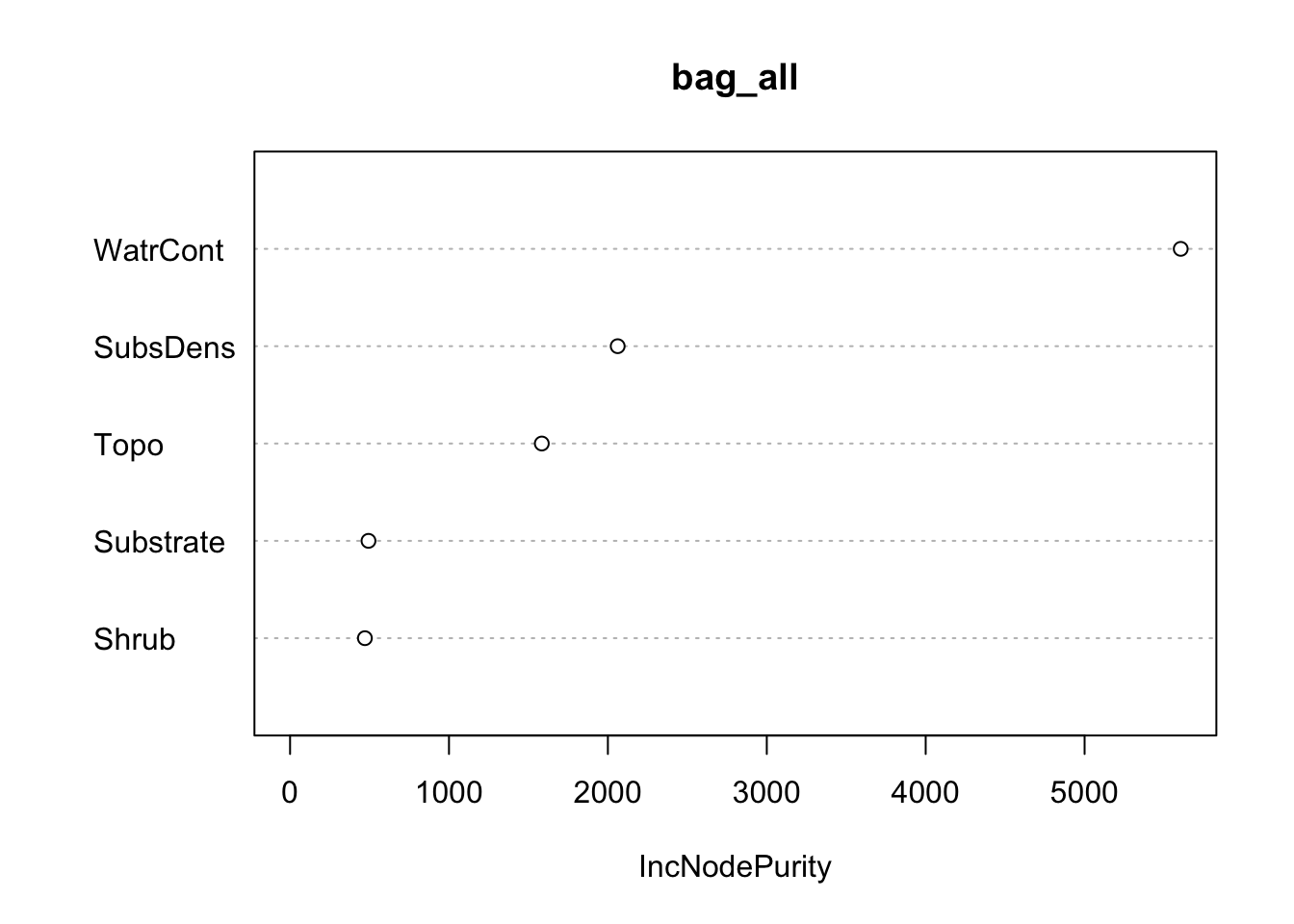
In order to obtain a measure of how “important” each predictor is by accessing the importance component. For regression tasks, this corresponds to the total amount that MSE decreases due to splits over a predictor, averaged over B:
IncNodePurity
SubsDens 2061.6646
WatrCont 5604.9744
Substrate 493.4425
Shrub 470.8712
Topo 1584.0990 IncNodePurity
SubsDens 2061.6646
WatrCont 5604.9744
Substrate 493.4425
Shrub 470.8712
Topo 1584.0990We can use the varImpPlot() function to visualize the importance:
Random Forests
The syntax for random forests is almost identical to that of bagging regression trees. Unlike bagging, we need to specify two parameters for random forests:
- The number of bootstrap samples \(B\) to obtain (as in bagging)
- The number of predictors we should consider at each split (i.e. the
mtryargument)
In the following code, I fit B = 10 regression trees, and specify mtry = 2.
Everything else is exactly the same as in bagged regression trees!